Research objectives:
What is the impact of post-transcriptional and post-translational processing of RNA and proteins on their interactions and location?
What are the changes in interactions/location upon perturbation, for example during drug treatment?
Do proteins adopt different structures/functions in different locations (moonlighting)?
What role does RNA play in moonlighting (for example RNA-protein interactions and/or localized translation)?
What is the spatial relationship between a protein and its corresponding mRNA?
To enable us to answer the above questions, we have developed a number of technologies and computational tools:
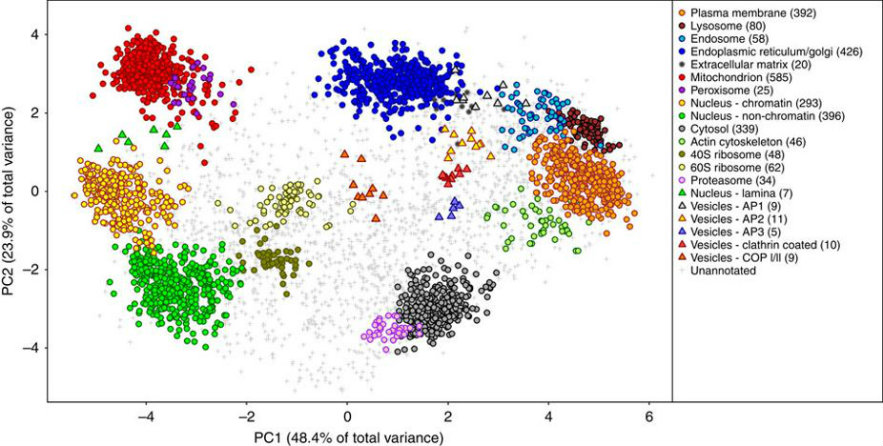
1. Spatial Proteomics
LOPIT - localization of organelle proteomics using isotope tagging
Geladaki A, Kočevar Britovšek N, Breckels LM, Smith TS, Vennard OL, Mulvey CM, Crook OM, Gatto L, Lilley KS (2019). Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics. Nat. Commun., 10(1):331. doi: 10.1038/s41467-018-08191-w
2. RNA binding proteome
OOPS - Orthogonal Organic Phase Separation

Villanueva E, Smith T, Queiroz RML, Monti M, Pizzinga M, Elzek M, Dezi V, Harvey RF, Ramakrishna M, Willis AE, Lilley KS (2019). Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol., 37(2):169-178. doi: 10.1038/s41587-018-0001-2
Queiroz RML, Smith T, Villanueva E, Marti-Solano M, Monti M, Pizzinga M, Mirea DM, Ramakrishna M, Harvey RF, Dezi V, Thomas GH, Willis AE, Lilley KS (2020) Efficient recovery of the RNA-bound proteome and protein-bound transcriptome using phase separation (OOPS). Nat. Protocols doi:10.1038/s41596-020-0344-2
3. Biotin painting to reveal in vivo structures
Minde DP, Ramakrishna M, Lilley KS (2020). Biotin proximity tagging favours unfolded proteins and enables the study of intrinsically disordered regions. Commun. Biol., 3(1):38. doi: 10.1038/s42003-020-0758-y
4. Protein protein interactions
Rees, J. S.; Lowe, N.; Armean, I. M.; Roote, J.; Johnson, G.; Drummond, E.; Spriggs, H.; Ryder, E.; Russell, S.; St Johnston, D.; and Lilley, K. S. In vivo analysis of proteomes and interactomes using Parallel Affinity Capture (iPAC) coupled to mass spectrometry. Mol. Cell Proteomics, 10(6):M110.002386
5. Computational Proteomics
Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M (2020). DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods, 17(1):41-44. doi: 10.1038/s41592-019-0638-x
Crook OM, Mulvey CM, Kirk PDW, Lilley KS, Gatto L (2018). A Bayesian mixture modelling approach for spatial proteomics. PLoS Comput. Biol., 14(11):e1006516. doi: 10.1371/journal.pcbi.1006516
New Horizons for CCP
1. CCP moves into the Milner Therapeutics Institute - October 2019

2. The Lilley group has recently started a project with the Theodoulou group (Rothamsted Research) the Mott group (UCL Genetics Institute) funded by a BBSRc sLoLa award to study 'What determines protein abundance in plants'
3. COVID-19 project with Ralser lab in Berlin
Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection.Messner CB, Demichev V, ... Ralser M.Cell Syst. 2020 Jun 2:S2405-4712(20)30197-6. doi: 10.1016/j.cels.2020.05.012.